A team of researchers at Nagoya University in Japan has created a 3-Dimensional computational simulation of the process of genome structure formation in the human cell nucleus. They expect the model to contribute to the understanding of cellular regulatory mechanisms and diseases, such as cancer, that damage the genome.
The three-dimensional structure of the genome plays a vital role in regulating the DNA functions of animal and plant cells because it affects its reading and replication. Shin Fujishiro and Masaki Sasai, Professor Emeritus of Nagoya University’s Department of Complex Systems Science, Graduate School of Informatics, constructed a 3D model by analyzing the whole genome of human cells. They used this model as a platform to investigate the relationship between the structure, dynamics, and functions of the human genome. The results were reported in the online edition of the Proceedings of the National Academy of Sciences of the United States of America.
“The spatial organization of DNA and its dynamic movement in cells are crucial for understanding cell functions,” Professor Sasai explains. “Researchers have devoted considerable effort to explaining DNA organization in cells by developing various experimental methods, including biochemical and microscope technologies, but now a unified picture is required. Our research introduces the first computational model that can quantitatively analyze various data from the full genome of human cells in a consolidated way.”
To help understand the process, the researchers investigated chromatin. Chromatin is a mixture of DNA with proteins that exist in cells as a way to keep the DNA compact. According to their model, chromatin is unevenly distributed and during the unfolding process that occurs during cell division, repulsive forces among the chromatin chains induce them to separate. This same force also separates chromatin into active and inactive compartments in the nuclei. The researchers found that their proposed mechanism clarified biochemical and microscopic findings of previous studies.
Professor Sasai adds: “Our model provides an indispensable tool and an original perspective in cell biology. From the computational model developed in this research, we can determine how perturbations in cells affect genome dynamics and organization. We can also investigate how disease cells, including various cancer cells, affect the genome. It allows us to more deeply analyze the relationship between genome structure and transcription regulation. The model developed in this research provides a fundamental tool and a new viewpoint in cell biology.”
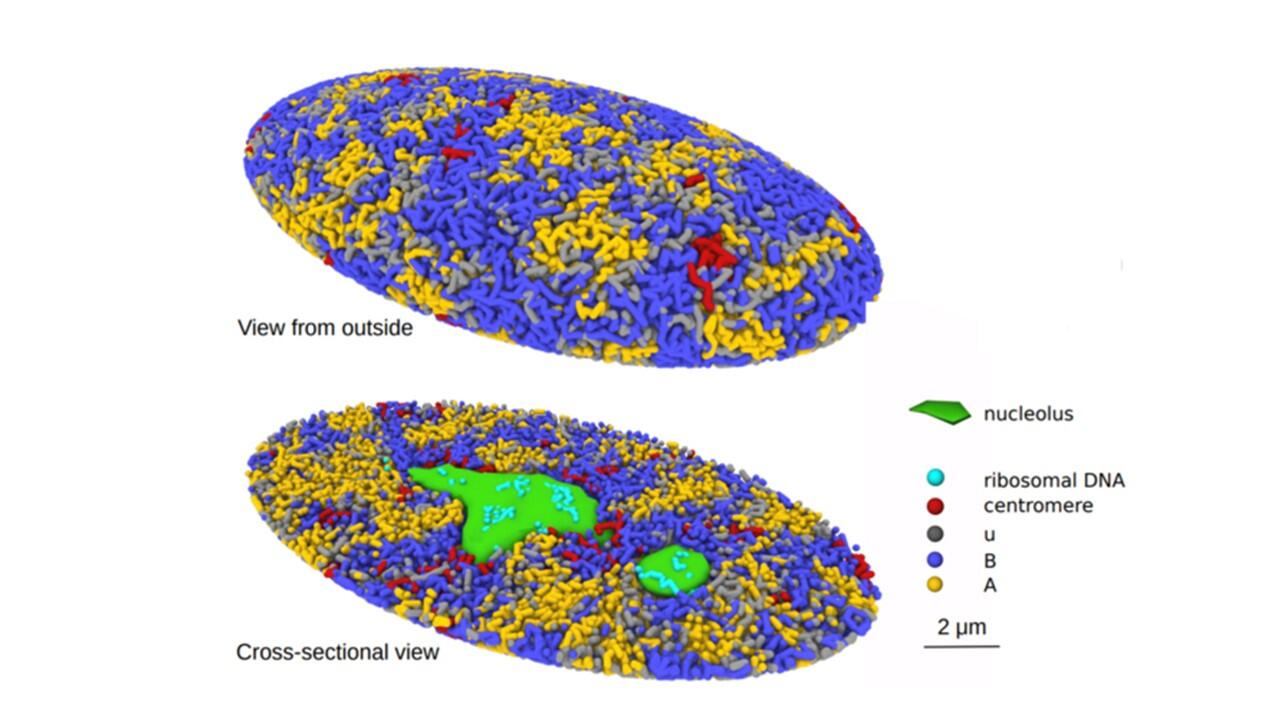
The study, “Generation of dynamic three-dimensional genome structure through phase separation of chromatin,” was published in the journal Proceedings of the National Academy of Sciences of the United States of America on May 26, 2022, at DOI: 10.1073/pnas.2109838119
Authors
Shin Fujishiro and Masaki Sasai, Nagoya University
This research was supported by the Japan Science and Technology Agency, Core Research for Evolutional Science and Technology (CREST) (JPMJCR15G2), RIKEN Pioneering Project, Grant-in-Aid for Scientific Research on Innovative Areas "Chromatin Potential" (19H05258, 21H00248), Grant-in-Aid for Scientific Research on Innovative Areas "Information Physics of Life" ( 20H05530), Grant-in-Aid for Scientific Research on Innovative Areas B (19H01860), and Grant-in-Aid for Scientific Research on Innovative Areas A (22H00406) supported by the Nagoya University Research Fund.
Media Contact
Matthew Coslett
- International Communications Office, Nagoya University
- kouho-en@adm.nagoya-u.ac.jp